Zorya anti-bacteriophage defense system ZorAB, ZorA delta_359-592, ZorA tail middle deletion.
Hu, H., Taylor, N.M.I.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Anti-phage defense ZorAB system ZorA | 495 | Escherichia coli | Mutation(s): 0 Gene Names: zorA, AW119_26345, BF481_003653, CQ986_005198, GAJ26_23520 | 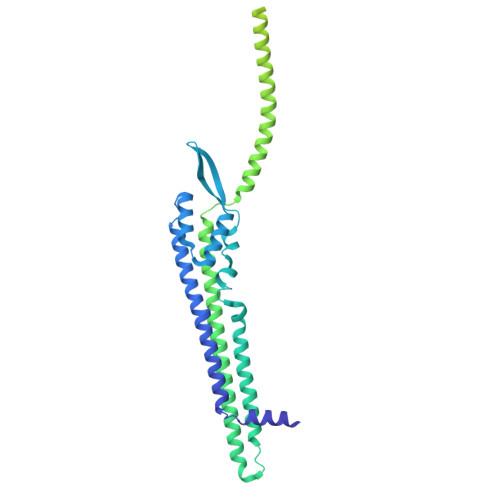 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Membrane protein | 246 | Escherichia coli | Mutation(s): 0 Gene Names: AW119_26350 | 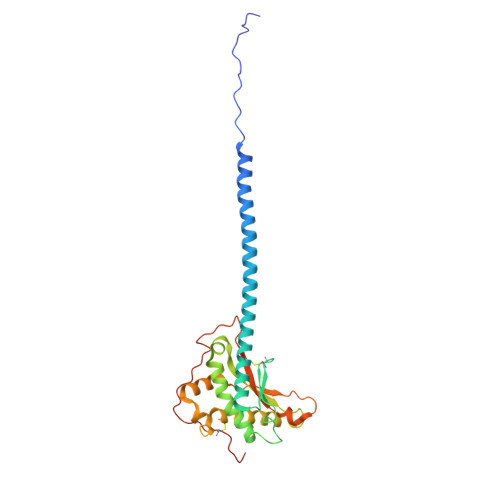 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CDL (Subject of Investigation/LOI) Query on CDL | H [auth A], K [auth B], N [auth C], Q [auth D], T [auth E] | CARDIOLIPIN C81 H156 O17 P2 XVTUQDWPJJBEHJ-KZCWQMDCSA-L |  | ||
| PEE Query on PEE | I [auth A], L [auth B], O [auth C], R [auth D], U [auth E] | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine C41 H78 N O8 P MWRBNPKJOOWZPW-NYVOMTAGSA-N |  | ||
| CA Query on CA | J [auth A], M [auth B], P [auth C], S [auth D], V [auth E] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Novo Nordisk Foundation | Denmark | NNF17OC0031006 |
| Novo Nordisk Foundation | Denmark | NNF21OC0071948 |
| Lundbeckfonden | Denmark | R347-2020-2429 |