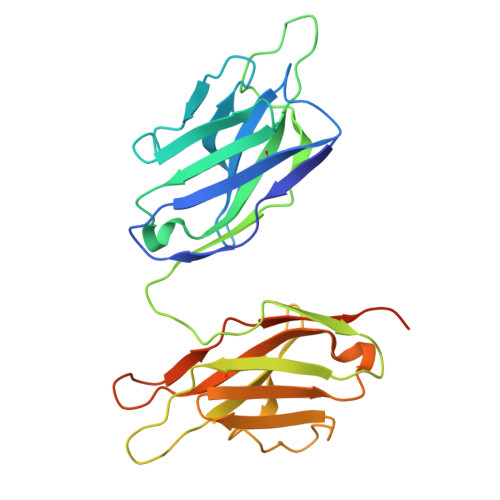
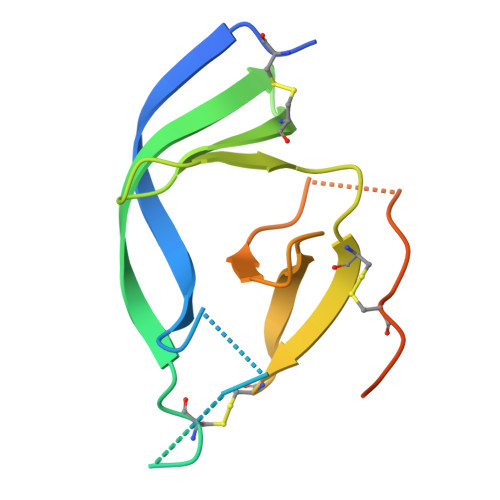
Structural insight into rabies virus neutralization revealed by an engineered antibody scaffold.
Kedari, A., Iheozor-Ejiofor, R., Salminen, P., Ugurlu, H., Makela, A.R., Levanov, L., Vapalahti, O., Hytonen, V.P., Saksela, K., Rissanen, I.(2024) Structure
- PubMed: 39471803
- DOI: https://doi.org/10.1016/j.str.2024.10.002
- Primary Citation of Related Structures:
8R3W, 8R40 - PubMed Abstract:
Host-cell entry of the highly pathogenic rabies virus (RABV) is mediated by glycoprotein (G) spikes, which also comprise the primary target for the humoral immune response. RABV glycoprotein (RABV-G) displays several antigenic sites that are targeted by neutralizing monoclonal antibodies (mAbs). In this study, we determined the epitope of a potently neutralizing human mAb, CR57, which we engineered into a diabody format to facilitate crystallization. We report the crystal structure of the CR57 diabody alone at 2.38 Å resolution, and in complex with RABV-G domain III at 2.70 Å resolution. The CR57-RABV-G structure reveals critical interactions at the antigen interface, which target the conserved "KLCGVL" peptide and residues proximal to it on RABV-G. Structural analysis combined with a cell-cell fusion assay demonstrates that CR57 effectively inhibits RABV-G-mediated fusion by obstructing the fusogenic transitions of the spike protein. Altogether, this investigation provides a structural perspective on RABV inhibition by a potently neutralizing human antibody.
Organizational Affiliation:
Institute of Biotechnology, Helsinki Institute of Life Science (HiLIFE), University of Helsinki, Helsinki, Finland.